
De Bruijn Graph of a Palindromic Sequence
Palindromic nucleotide sequences are those matching their reverse complements. Here is an example -“ATACGTCATTCAAATATGTATATACATACATATTCGAATATGTATGTATATACATATTTGAATGACGTAT”. Another one is “CATGTGACTTTATAGCAGCATGCTGCTATAAAGTCACATG”. We did not make those sequences up. They are taken from chromosome 1 of human genome.
Here is the de Bruijn graph of one of the above sequences with 7-mers. We are using the convention of showing only the lexicographically smaller 7-mer.
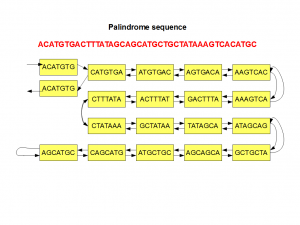
The above graph is constructed in a hurry and may have one or two errors, even though the general structure is correct.
Enjoy !!