
Long Term Direction of Nanopore Sequencing - Three Ways from Here
An infinite amount of propaganda being spread about Oxford Nanopore really troubles the lowly janitors like us. We are not sure why this company and its ‘fanboys’ operate with innuendos passed around social media channels and not deliver any real information like other respectable companies would do. Hopefully Nick Loman’s presentation tomorrow will be backed by release of some real data, but until then scientists’ job is not to cheer-lead for companies, but find out the truth and represent it faithfully and accurately. The scientific community is failing to play its proper role just like they failed to debunk Ewan Birney’s misleading media campaign about ‘killing the junk DNA’ (@ENCODE_NIH). In fact, in case of ENCODE, scientists were so married to the propaganda from Birney and friends that even reputed journals attacked Dan Graur for simply telling the truth.
We sincerely hope that the ONT saga is not a repeat of that experience (despite the involvement of disgraced scientist Ewan Birney in both), but let us deconstruct some of the myths surrounding nanopore sequencing.
-————————————–
Myth 1. “Oxford Nanopore is a small company.”
BioMickWatson is trying to spread this myth through his blog post posted over the weekend.
According to Bloomberg news, the company had a valuation of $1.5B last year and the valuation must have gone up during last month’s fundraising.
The company was spun out of Oxford University in 2005 and its technology was initially based on research by Hagan Bayley, a professor of chemical biology at the university. Numis Securities analyst Charles Weston estimated the companys value at $1.5 billion in a Feb. 20 note to clients.
A $1.5B-$2B company is anything but small. For comparison, PacBio’s current market cap is only ~$386M. By another measure, ONT raised ~$300M over the years. That is not pocket change except to those working on ENCODE project.
In fact, it is quite likely that being big (in terms of valuation) is ONT’s real problem. The company is priced for perfection and has to execute flawlessly. That is quite stressful for its managers as we can imagine, but it is not an excuse for the scientific community to shy away from its responsibilities.
-————————————–
Myth 2. “The company is highly secretive.”
This one was being passed around last year, when one minute of google search could show that the company disclosed almost everything it was doing in its patent applications. Those applications were all part of public information.
Everything You Want to Know about Oxford Nanopore
Any curious soul could easily find out the above information, but the ‘highly secretive’ meme continued.
-————————————–
Myth 3. “ONTs stated philosophy is for customers to control and release data (as is happening).”
This whopper was dealt to us by a reader of BioMickWatson’s blog, who appeared knowledgeable about ONT’s strategies. In fact, if there is anything the company is secretive about, it is in releasing any data. For example, check a post from OmicsOmics blog in Feb 2012 -
Why Oxford Nanopore Needs to Release Some Data Pronto (Besides Bailing Me Out)
Last week’s piece on Oxford Nanopore got a lot of attention and a lot of comments, which to me is the true mark of success in this space (discounting the higher than normal spam attempts). A couple of folks were kind enough to tweet a link (now captured in Nick Loman’s wonderful tweet archives for AGBT 2012), and it was also picked up by Matthew Herper at Forbes, Dan Kobolt at MassGenomics and others (apologies for all I haven’t shouted out). It also can’t be denied that some of those comments felt I had been too generous / gullible with Oxford Nanopore. It’s always a trick writing pieces about new technologies, and I’m aware of a number of factors which can influence my take on a given story.
Or Lex Nederbragt’s comment from Oct 2013 -
Would you buy that washing machine?
Ever since the first announcement of the MinIon and GridIon, the community has been asking for, and becoming more and more frustrated with, the lack of real Oxford Nanopore sequencing data to look at. Most sequencing companies (admittedly, not all) happily share their latest and greatest reads for the community to tear apart. But Oxford Nanopore just trusts us to believe the hype and buy into this early access program. Reports on twitter indicate some researchers and even a bioinformatician were able to run the MinIon at ASHG. But still NO DATA was available after these runs.
In fact, we can deconstruct the myth that “ONTs stated philosophy is for customers to control and release data (as is happening)” in one minute. Would David Jaffe please release his data from his AGBT talk or explain the reason for not doing so? That talk made several specific claims about the technology (see Nanopore talk of David Jaffe) and we do not see how holding on to the data benefits the scientific community. Dr Jaffe, would you please let us evaluate the claims you made in your AGBT talk?
-————————————–
Myth 4. “ONT error rate - better than pacbio but worse than Illumina.”
This was spread around last year’s ASHG. For example, please check the following comment by Heng Li in Yaniv Erlich’s blog. Heng is a respectable scientist and we have no reason to believe he makes up claims unless he hears from ONT booth at the conference or other insiders.
The numbers I heard from ASHG: tens of megabases per hour for 6-8 hours (similar to your number). The error rate is better than pacbio but worse than Illumina. Perhaps harder in tandem repeat (if I understand correctly). Read length is “tens of kb”. Look forward to it.
The above myth got deconstructed this week, when ONT fanboys claimed that neither BLAST nor BLASR was good for aligning nanopore reads. Hmmm….less error rate than PacBio, but BLAST does not work?
-————————————–
Myth 5. “Nick Loman already released nanopore data.”
BIO-IT world reported -
First Reads from Nanopore Sequencers Becoming Public - By Bio-IT World Staff
This morning, a trio of U.K. researchers released what may be the first publicly available read from an Oxford Nanopore MinION Sequencer. Oxford Nanopore opened their early access program for the thumb drive-sized sequencers in November, and began distributing instruments earlier this year.
Nicholas Loman, Josh Quick, and Szymon Calus, who have previously collaborated on molecular diagnostics for infectious diseases, posted their MinION data on the data-sharing site figshare.
NextGenSeek wrote -
Behold. Oxford Nanopore Reads Are Here
We would like to inform the staff of Bio-IT World that they have been bamboozled, because the ‘trio’ released exactly only one read through Figshare and not even two to make their title valid. This is a complete joke and waste of everyone’s time, when some data should have been released by David Jaffe six months back. Apparently, BioMickWatson himself also got fooled by the same media reports (check his comment).
We again heard that there are plans to release some data this week, when Nick Loman and Oxford Nanopore CEO Clive Brown present on nanopore technology. Keith Bradnam wrote -
The data should be made available this week. These are very large datasets and Nick is taking the steps to release such data in an appropriate manner.
Let us hope so, but until then Mikheyev’s released data are the only publicly available ones for scientists to rely on.
-————————————–
Myth 5. “Mikheyev used old chemistry, which improved since then.”
Check - Oxford Takes Some Flak, Fires Back
Benchmarks are a good thing, but it can be strongly argued that this one was grossly rushed and premature. In particular, while the article doesn’t state the chemistry used, the timing on the paper clearly shows this work was performed using the initial shipment of flowcells and kits. This was a chemistry that ONT was open about: not their best, but it was ready-to-ship. The second set of flowcells delivered their next chemistry, and the next set will have a tweak on that. Oxford is rapidly iterating on every aspect of their platform: library preparation, protocols, flowcells and base callers, but this paper is based on the very first version.
We have no reason to believe that a early-stage (started in 2005) sequencing company cannot greatly improve its chemistry over time, but David Jaffe made the following claim in his AGBT talk prior to MAP program -
84% of 5+kb reads have at least a perfect 50-mer, 100% have at least a perfect 25-mer
Why did not Oxford Nanopore give access to at least the same sequencing technology to Mikheyev and alike ?
-————————————–
Myth 7: Our blog instructed Mikheyev to publish his paper on ONT.
Readers may have seen this comment posted at our blog two days back.
Hey samanta, I know you are a massive advocate of open science. Maybe you could publish your correspondences with Mikheyev over the last 6 months ?
These nanopore fanboys are so out of their mind that they think the world lacks scientists thinking independently. We never knew about Mikheyev prior to last week, but let us create some hypothetical conversation to make this reader happy.
A series of ‘hypothetical’ emails exchanged between us over six months:
Mikheyev to Homolog.us: “Hey, we got a nanopore sequencer as part of the MAP early access program.”
Homolog.us to Mikheyev: “Great!! Let us do something nefarious with it.”
Mikheyev to Homolog.us: “Nefarious like what?”
Homolog.us to Mikheyev: “…like telling the truth. Note - do not leave Japan, because telling the truth has been declared a terrorist act in UK. Secondly, you must not publish in glam journals and do make sure you release all your raw data publicly.”
Mikheyev to Homolog.us: “I don’t get that. Won’t people discredit our report, if we do not publish in Science or Nature?”
Homolog.us to Mikheyev: “Scientific world has changed and become very open minded about where you publish. Listen to what the influential people are saying these days. The journal does not matter as long as you do proper science and publicly release all your data to back the claims.”
Mikheyev to Homolog.us: “Should we report, if our results are negative?”
Homolog.us to Mikheyev: “Report whatever you believe is the truth.”
We made up the conversation, but hope it satisfies the reader.
-——————————————
Myth 8: Scientists need to stay positive about nanopore technology due to its disruptive nature.
This is such a large whopper that its sponsors can probably buy Burger King, now that the company is up for sale.

What do we do next - tilt clinical trials toward ‘positive’ side, because drug companies are working on disruptive and groundbreaking technologies? Offer Nobel prize to the positivity lady for her outstandingly dubious science? Play the stupid ‘Happy’ song five times a day, just because the lyrics say happy, happy, happy? When did the notion of objectively reporting based on available evidence go out of fashion?
As we see, nanopore sequencing can take three ways from here, and scientists need to judge each possibility and report truthfully without innuendos and disinformation. Those three ways are -
(i) The early access program shows that ONT’s nanopore technology works just promised and the company becomes immensely successful. That will definitely benefit scientific communities.
(ii) Despite huge potential for being disruptive, nanopore sequencing never flies. The technology behind flying cars would be a good comparison.
(iii) Nanopore technology works after several trials and errors and takes time to get adopted until all its flaws are worked out and proper bioinformatics tools are developed. That is quite a respectable solution for the scientists, even though it does not support the high valuation of the company. The company may even get sold to Chinese for low price like a number of companies mentioned in this report.
The MIT Curse Strikes Again: Lilliputian Systems Files For Bankruptcy
Everyone writing about ‘being positive’ about the company and the technology do not take this third possibility into account, even though that had been the course for most disruptive technologies. Handheld computer like Ipad did not come overnight and Palm, Handspring and a number of almost successful companies went away. Here is how things played out for the mother of all disruptive technologies, its inventor and the city that backed him.
Johann Gutenberg (c. 1398-1468), the German printer, is supposed to have been born c.1398-1399 at Mainz of well-to-do parents, his father being Friele zum Gensfleisch and his mother Elsgen Wyrich (or, from her birthplace, zu Gutenberg, the name he adopted). He is assumed to be mentioned under the name of Henchen in a copy of a document of 1420, and again in a document of c. 1427-1428, but it is not stated where he then resided. In 1420 the citizens of Mainz drove the patricians out of the city in a tax revolt, and as Gutenbergs name appears about ten years later at Strasburg, the family probably took refuge there.
A new city government emerged established by the tax revolt forbade the sale of any more annuities without the consent of the trade guilds. Despite this tax rebellion between the trade guilds and the patricians, the citys financial situation continued to decline as it effective sent the rich fleeing. Clearly, with the rich gone, the city was unable to revive its economy having effectively destroyed Adam Smiths Invisible Hand. This led to the expelled families being recalled to Mainz, for we now see that Gutenberg did not avail himself of the privilege since he is described in the Act of Reconciliation (dated March 28) as not being in Mainz. The return of the patricians may have been predicated upon their buying debt of the city since on January 16, 1430, Gutenbergs mother arranged with the city of Mainz to purchase an annuity belonging to her son. This appears to be the reason for the recall of the expelled rich when the city cannot revive its economy without them.
We next hear of Gutenberg at Strasburg, where he was associated with the goldsmiths guild there between 1434 and 1444. In 1434 he seized and imprisoned the town clerk of Mainz for a debt due by the corporation of that city. However, he released him at the urgent representation of the mayor and counselors of Strasburg, and he relinquished at the same time all claims to the money (310 Rhenish guilders = about 2400 mark silver). In 1437 Gutenberg was sued before the ecclesiastical court by Emmeline zu IserneThure for breach of promise of marriage, but the case was settled by his making the woman his wife. In 1442 he borrowed 80 from the Chapter of Saint Thomas in Strasbourg that he appears never paid back. He seems to have returned to Mainz around 1444, since we do know in 1448 on October 6th, he borrowed 150 gulden. It was two years later when he borrowed 800 gulden from Johann Fust, a lawyer and wealthy merchant banker. In 1452, he borrowed another 800 gulden from Fust making him a partner. This was an incredible amount of money equal to about 10-years wages.
Finally, in 1436-1437, 75% of the total expenditure of Mainz went to creditors, whose interest payments continued to increase crowding out all economic growth. The interest expenditures were draining the economic fortunes of Mainz and now there was an ever increasing difficulty to find new subscribers to its loans. This escalated causing interest rates to rise. During the 1430s, Mainz offered 5% for the perpetual annuities instead of the previous 3% or 4%. The total national debt of Mainz reached 373,184 gulden. It was in 1448, when the city of Mainz could find no buyer of its debt and was unable to raise 21,000 gulden that it declared itself bankrupt. Since 60% of the debt was invested outside Mainz, the city was placed under Imperial ban, excommunicated by the Pope whereas today 40% of all interest paid by the United States goes overseas. Taxes were raised and the rich deserted the city further ensuring the collapse of its productive forces. With the richest bourgeois gone and the city deeply impoverished, the fortunes of the city turned negative.
Adolph II (or III) of Nassau-Wiesbaden-Idstein (German: Adolf II. von Nassau- Wisebaden-Idstein) (c. 1423 6 September 1475), was Archbishop of Mainz from 1461 until 1475. He was a son of Count Adolph II of Nassau-Wiesbaden-Idstein. On June 18, 1459 he was defeated in the election to the Archbishopric of Mainz by Theodoric of Isenburg-Bdingen who was never confirmed by the Pope. In 1461 he went to Nuremberg for Imperial and Papal reform, and its recommendations earned him the wrath of both the Emperor Frederick III and Pope Pius II. It was then in 1461 that Pope Pius II declared Adolph the Archbishop of Mainz following the confrontational reforms of Theodoric.
Since the city of Mainz and its and cathedral chapter remained loyal to Theodoric, Adolph declared war. The devastating Mainzer Feud continued for a year until on October 28th, 1462 Adolph captured the city. Some 400 citizens were executed, and another 400 fled abroad. Adolph also revoked Mainzs privileges and the status as an Imperial City. Mainz was sacked by Archbishop Adolph von Nassau, and Johann Gutenberg was exiled from Mainz and he moved Eltville where he may have initiated and supervised a new printing press belonging to the brothers Bechtermnze. Gutenberg died in 1468 and was buried in the Franciscan church at Mainz, his contributions largely unknown. This church and the cemetery were later destroyed, and Gutenbergs grave is lost.
-————————————————-
Myth 9: Scottish independence will be disruptive for businesses and scientists.
Unlike BioMickWatson, we see Scottish independence as one of the biggest positive developments of this decade. However, we can offer a plus that even he can appreciate. If science is redefined as cheer-leading for ENCODEsque big science and Ewan Birney’s companies, Scottish independence will provide appropriate clothing for the cheer leaders.
-——————————————
Lex Nederbragt wrote last year -
Is the MinIon Access Program more like The Emperors New Clothes, with all of us acting like the emperor?
Let us hope Nick Loman proves that wrong tomorrow so that we can objectively judge the merits of the technology. Even then, note that paths (i) and (ii) are both beneficial for scientists, and they should focus on science instead of trying to operate the business as well.
-————————————————————
Edit.
We are starting to receive a lot of real information on Nanopore. Readers are encouraged to check the long and helpful comment by David Eccles. Speaking of genome assembly, Michael Schatz wrote -
Michael Schatz ?@mike_schatz 16m
In case there was any question about de novo assembly with @nanopore. Here is yeast ~10x better than illumina alone
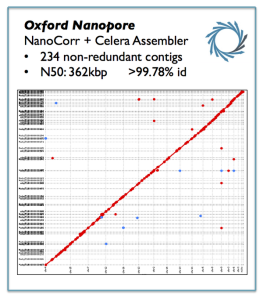
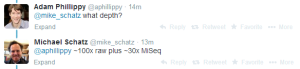
100X raw coverage means about 1.2 Gbp of sequences.