Using Cascading Bloom Filters to Improve the Memory Usage for de Brujin Graphs
Today’s must read paper appeared in arxiv (emphasis ours).
De Brujin graphs are widely used in bioinformatics for processing next- generation sequencing data. Due to a very large size of NGS datasets, it is essential to represent de Bruijn graphs compactly, and several approaches to this problem have been proposed recently. In this work, we show how to reduce the memory required by the algorithm of [3] that represents de Brujin graphs using Bloom filters. Our method requires 30% to 40% less memory with respect to the method of [3], with insignificant impact to construction time. At the same time, our experiments showed a better query time compared to [3]. This is, to our knowledge, the best practical representation for de Bruijn graphs.
Those unfamiliar with Minia can read our earlier commentaries.
Top Bioinformatics Contributions of 2012.
Quip, Minia, SlimGene and Titus Browns paper on Scaling Metagenome
Minia Assembler and French Revolution
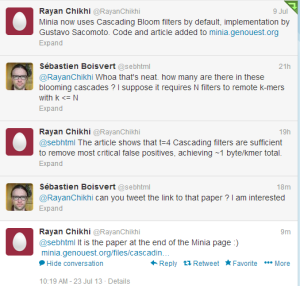
P.S. Adding a twitter conversation in case the same question comes to your mind, because five months later people will be chatting about different things in twitter.